I investigated the application of protein language models (PLMs) for immunoglobulin sequence analysis. Antibodies are adaptive immune proteins critical for pathogen neutralization. This study integrates B-cell receptor (BCR) sequencing data, AbLang (an antibody-specific protein language model), and single-cell RNA sequencing to explore computational approaches in immunoinformatics.
Protein Language Models in Structural Biology
Protein language models leverage transformer architectures adapted from natural language processing to analyze amino acid sequences. These models undergo unsupervised pre-training on large-scale protein databases, learning evolutionary constraints and structural motifs encoded in sequence space. Through self-attention mechanisms, they capture long-range amino acid dependencies critical for protein folding and function. The learned representations encode biophysical properties including secondary structure, solvent accessibility, and functional domains. I investigated their application to immunoglobulin evolution and somatic hypermutation analysis.
AbLang: Antibody-Specific Protein Language Model
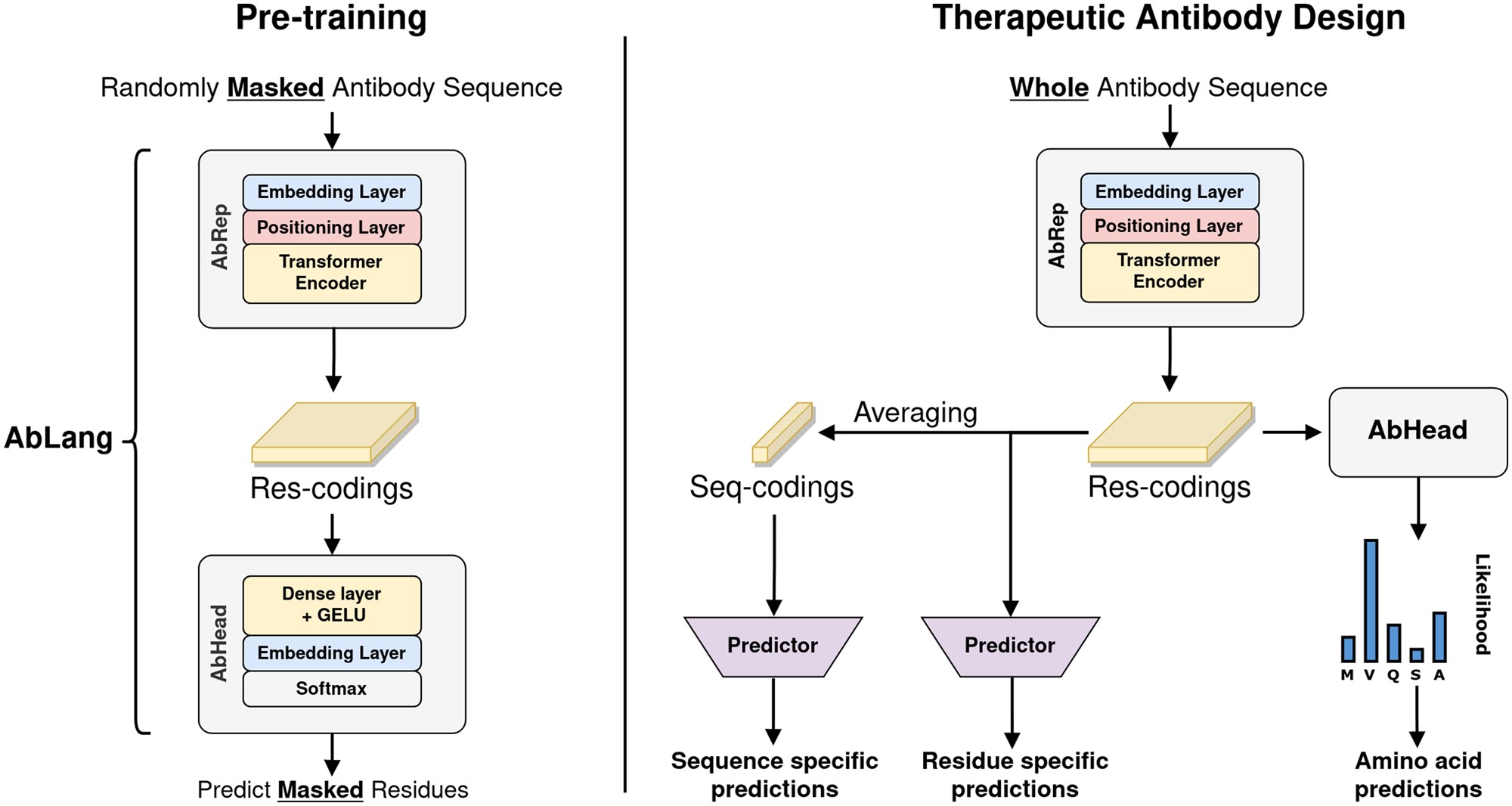
AbLang is a domain-specific protein language model trained exclusively on immunoglobulin sequences, optimizing representations for antibody variable regions. While domain-specific models may capture immunoglobulin-specific evolutionary patterns, comparative analysis with general protein language models (e.g., ESM-2, ProtTrans) would provide valuable benchmarking for immunoinformatics applications.
Additional Background about Antibodies at the bottom:
Somatic hypermutation frequency serves as a molecular marker of affinity maturation, with higher mutation loads indicating extensive germinal center selection and clonal evolution from naive B-cell precursors.
Overview of my investigation:
- integrate single-cell transcriptomic profiles with AbLang-derived immunoglobulin embeddings
- apply principal component analysis to protein language model representations
- perform unsupervised clustering of immunoglobulin sequence embeddings
- conduct differential gene expression analysis using embedding-defined B-cell subpopulations
- generate low-dimensional visualizations via UMAP projection
Protein Language Models for Immunoglobulin Analysis in Single-Cell Genomics
A Quick Look at the Dataset
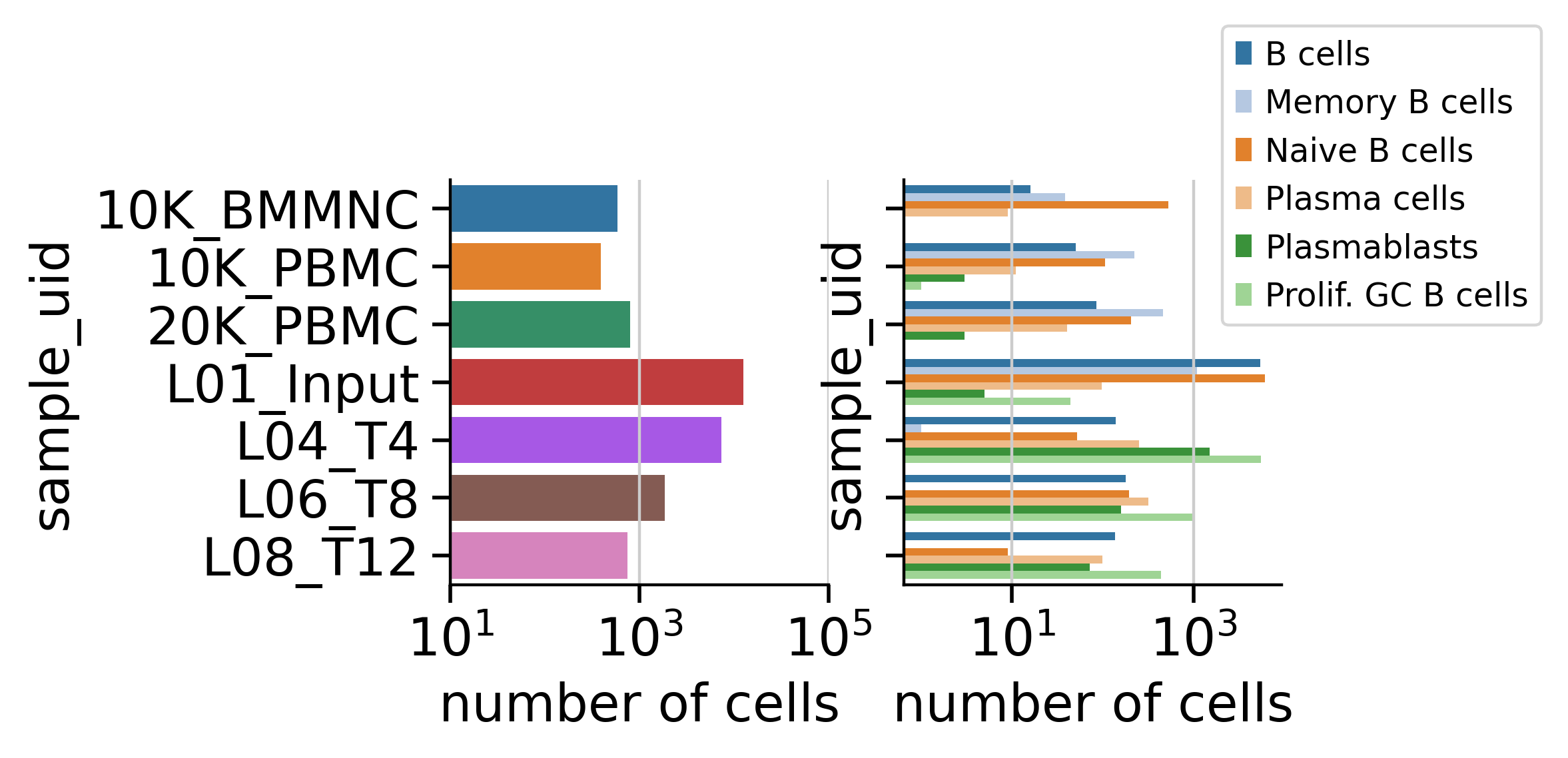
The dataset comprises hundreds to thousands of individual B cells per sample, each with paired single-cell transcriptomic profiles and cognate immunoglobulin heavy and light chain sequences.
These UMAP projections demonstrate global coherence between transcriptomic states and immunoglobulin sequence characteristics:
- UMAP projections of single-cell transcriptomic data colored by B-cell developmental state (left) and immunoglobulin sequence features (right panels)
- Memory B cells represent antigen-experienced populations with accumulated somatic hypermutations, while naive B cells maintain germline immunoglobulin sequences
- Mutation frequency is quantified as nucleotide divergence from inferred germline V(D)J gene segments
- Memory B cells exhibit class switch recombination from IgM to downstream isotypes (IgG, IgA, IgE), indicating germinal center participation
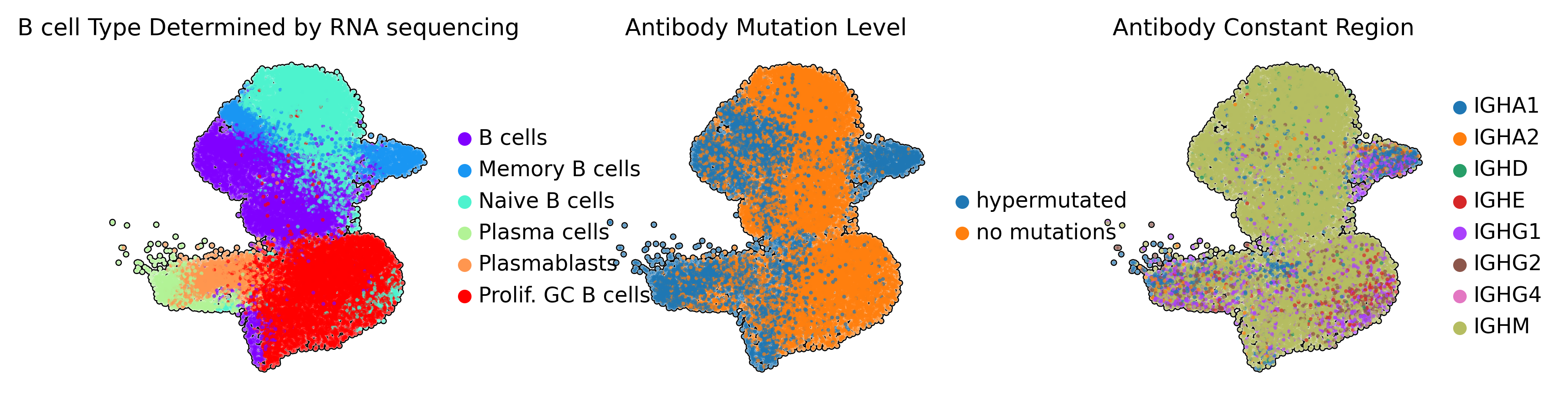
Immunoglobulin sequence annotations demonstrate strong concordance with transcriptomically-defined B-cell subsets. Memory B cells predominantly harbor hypermutated immunoglobulin sequences, confirming the molecular signatures of germinal center-derived populations.
I investigated whether protein language model embeddings capture biologically meaningful immunoglobulin features including somatic hypermutation and isotype usage. The following PCA analysis examines these categorical variables within AbLang embedding space:
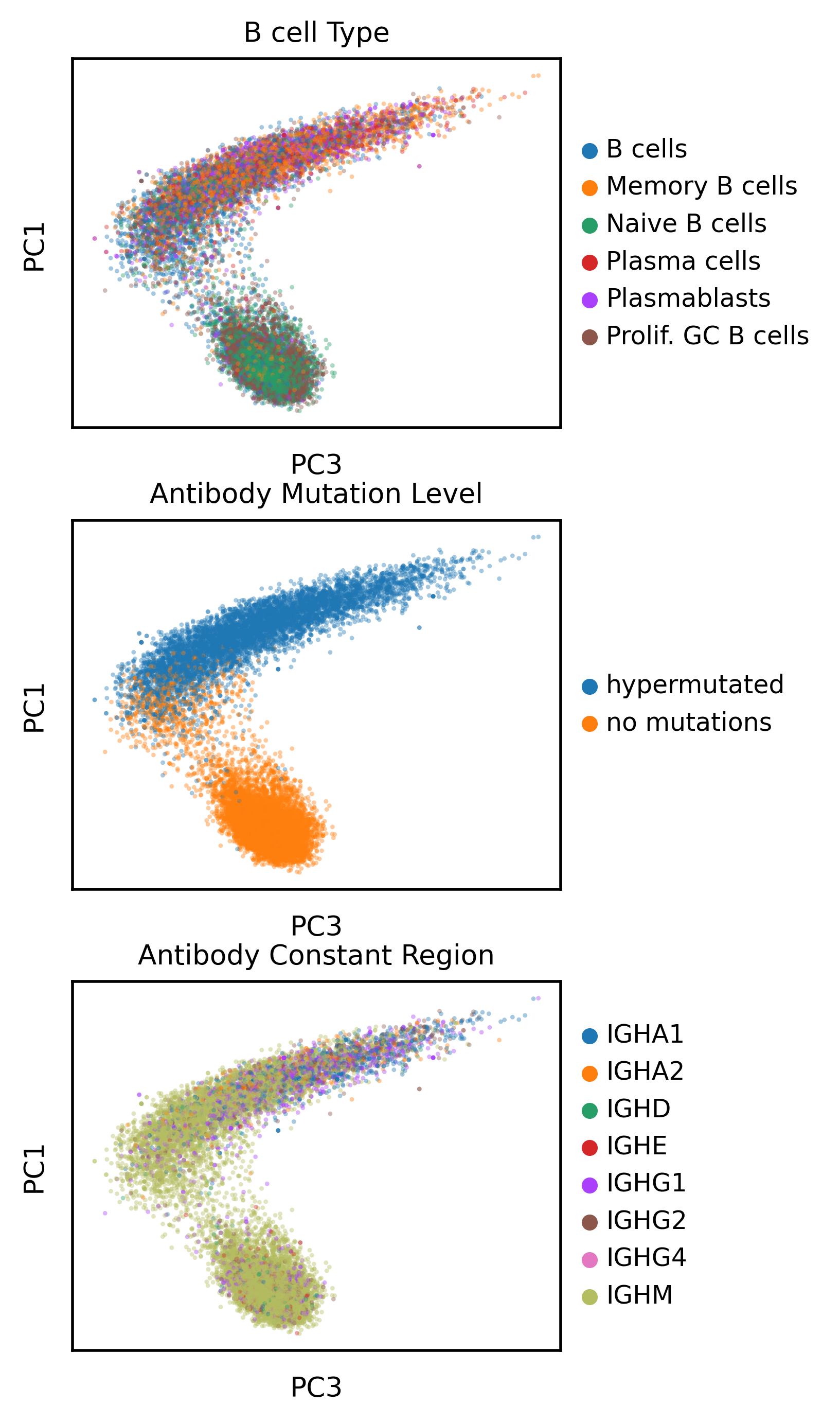
The protein language model demonstrates learned representations of somatic hypermutation, cleanly separating germline from hypermutated immunoglobulins along PC1 and PC3. PC3 reveals continuous variation within hypermutated sequence space, with a clear trajectory from IgM to class-switched isotypes (IgG, IgA). This suggests the model captures fundamental immunoglobulin biology including affinity maturation and class switch recombination.
Quantitative analysis of somatic hypermutation along principal components, particularly PC3 which separates IgM from class-switched isotypes, reveals corresponding patterns in transcriptomic space:
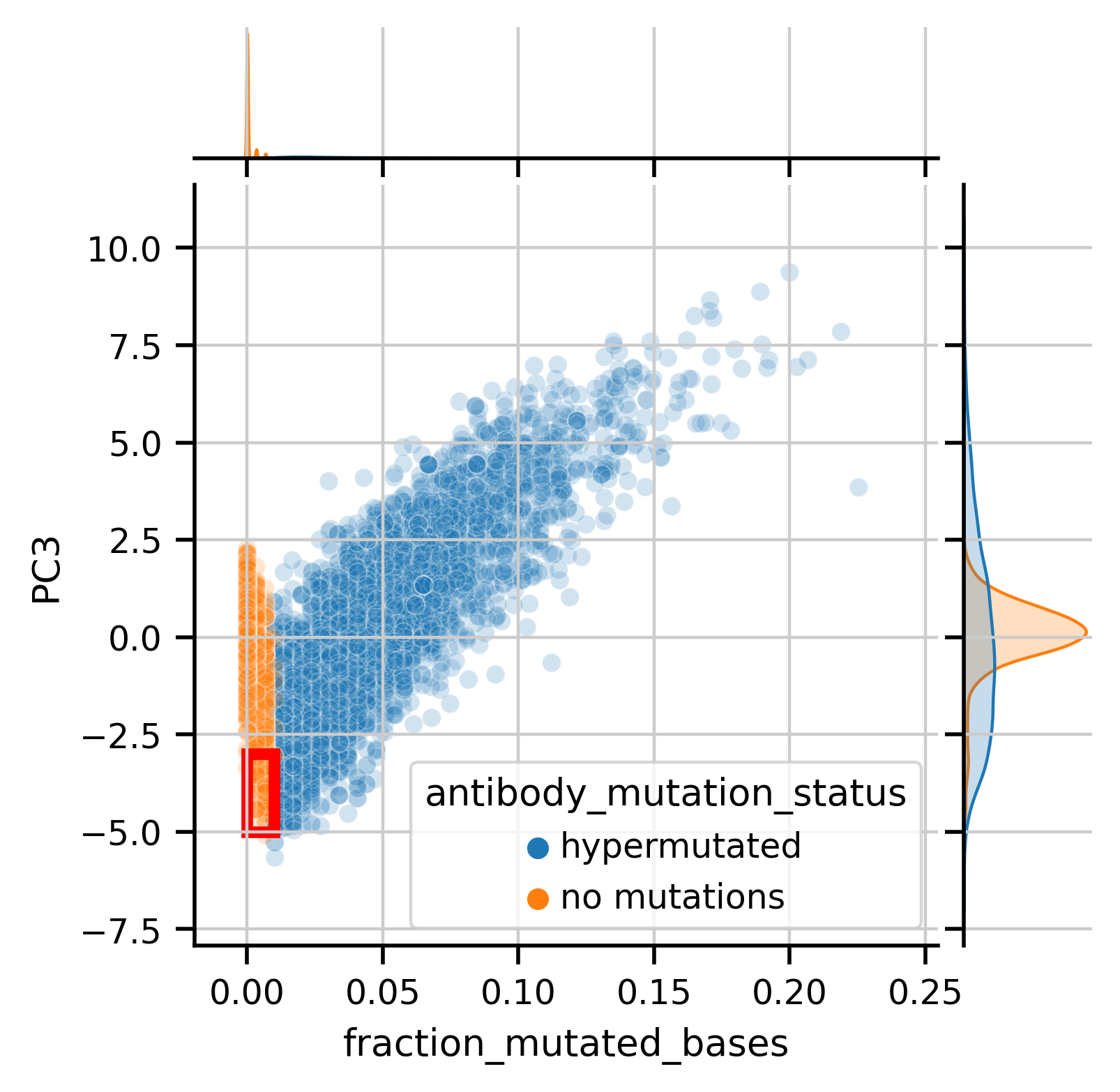
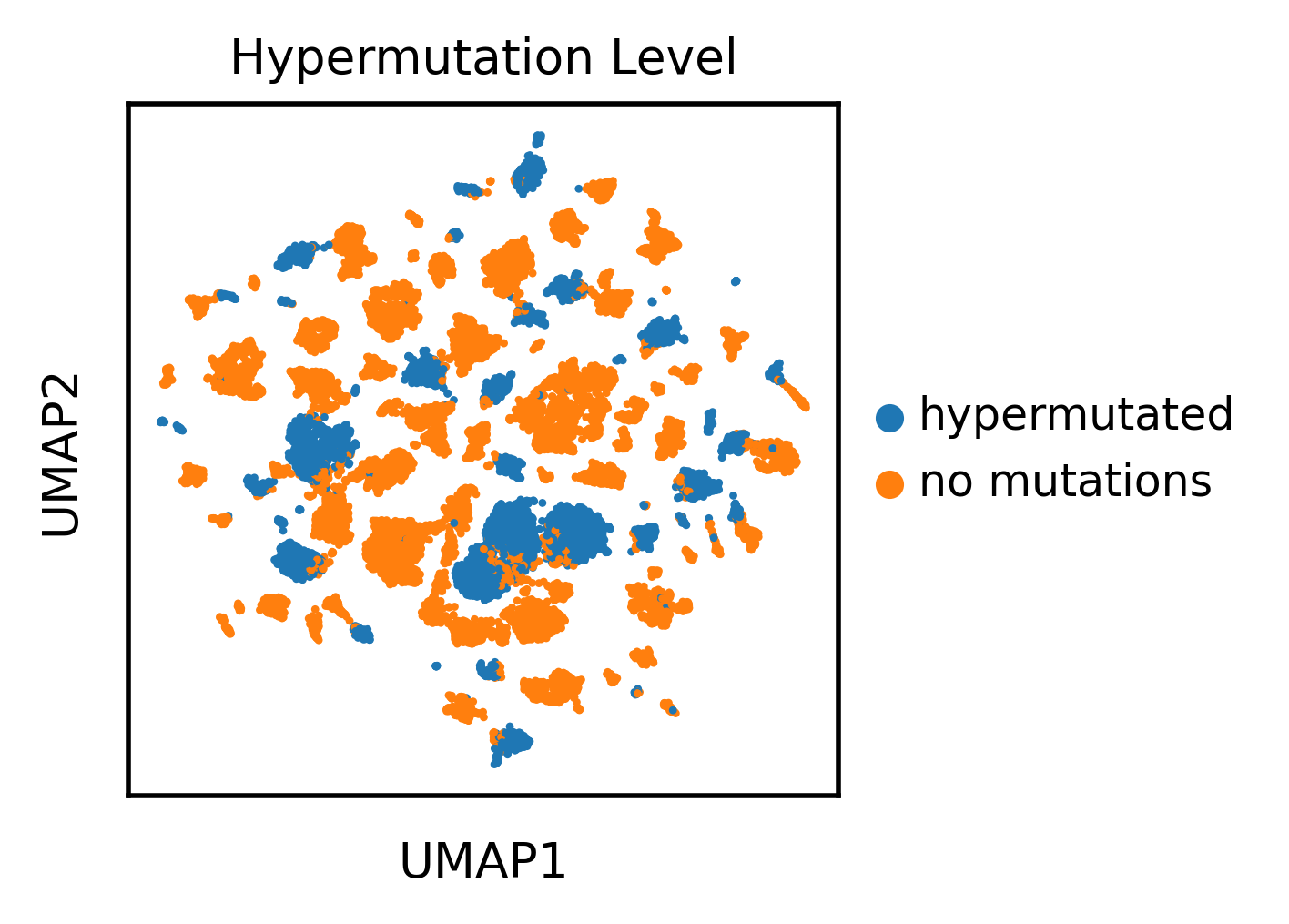
As an aside, we can see a fair amount of "no mutation" antibody sequences (highighted in the red box or seen in the tail of the orange distribtion) that actually appear more like mutated antibodies in the PC space. I asked if the gene expression of cells with that profile looked like Memory B cells, which would corroborate that the antibody actually is hypermutated. Indeed these cells with "no mutations" appear to have a memory-like phenotype, suggesting the language model identifies mutations in antibodies more sensitively than how I orginally labeled the mutation status, via comparison to a genomic database.
I noticed from some exploratory plots that the while the distribution of hypermutated antibodies on PC3 doesn't appear to have any notable features, if I subset different cell types, multiple modes appear. For example, hypermutated Memory B cells appear to have multple two modes in PC3
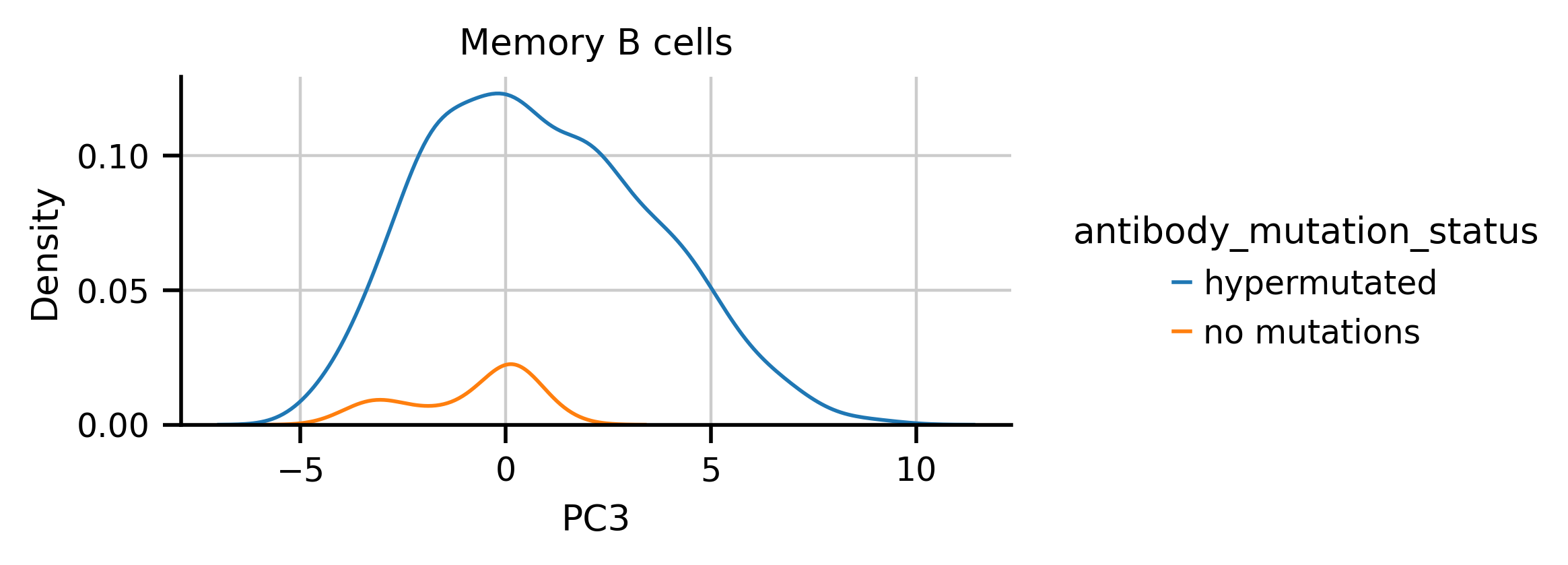
I split the hypermutated Memory B cells at 1 in PC3 space and asked if there were gene expression differences between the groups
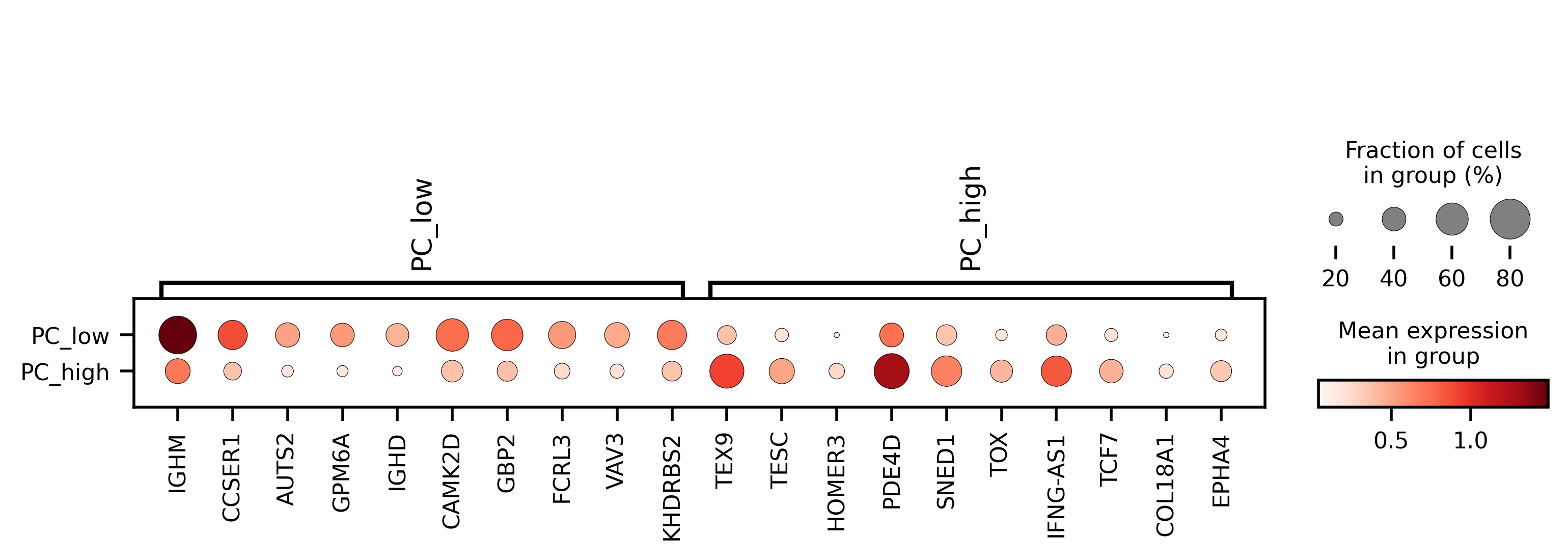
Indeed there are clear differences, with PC3 low having a IGHM memory phenotype and PC3 high deeper or class-switched memory phenotype
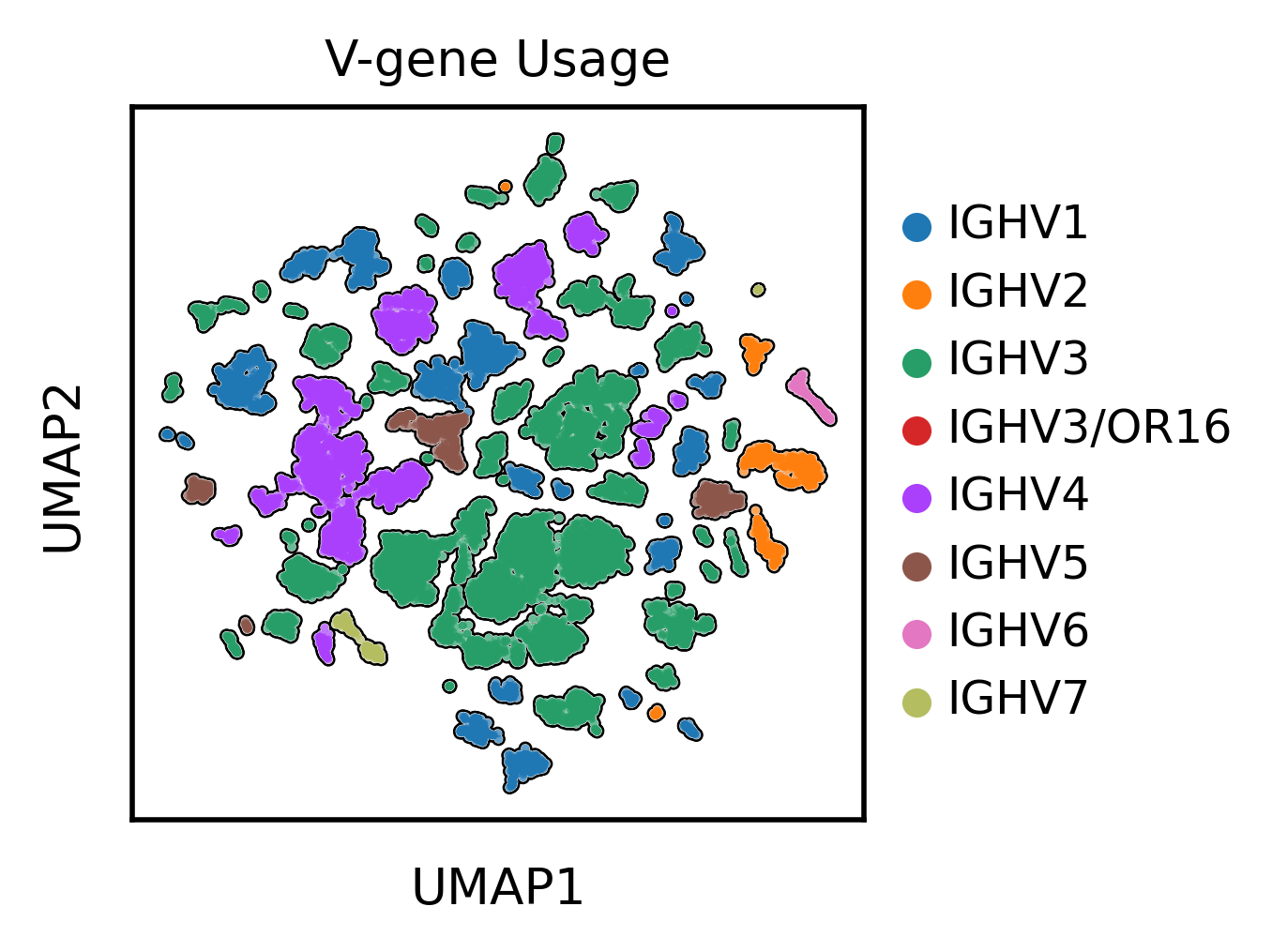
Let's finally take a quick look at the UMAP projection of the antibody sequence encodings

UMAP projection demonstrates clustering by V gene family usage alongside somatic hypermutation levels. Immunoglobulin sequences cluster by germline V gene origin, with secondary separation of hypermutated variants within each gene family, reflecting both germline diversity and somatic evolution.
Summary:
This immunoinformatics analysis demonstrates that protein language models capture biologically relevant immunoglobulin features:
- AbLang embeddings encode somatic hypermutation along orthogonal dimensions
- Binary germline vs. hypermutated classification along one axis
- Continuous mutation load quantification along another axis
- Embedding-based immunoglobulin clustering correlates with independent transcriptomic B-cell phenotypes
Future Directions:
- Quantify embedding space distances for clonally related immunoglobulins to assess phylogenetic signal preservation
- Develop cross-species protein language models incorporating mouse, camelid, and non-human primate immunoglobulin data for comparative immunogenomics
- Train memory-specific models exclusively on hypermutated sequences to enhance affinity maturation signal detection
Immunoglobulin Structure and Function
Immunoglobulins are tetrameric glycoproteins comprising two heavy chains and two light chains linked by disulfide bonds in a characteristic Y-shaped quaternary structure. The antigen-binding domain, formed by the variable heavy (VH) and variable light (VL) regions, determines specificity through complementarity-determining regions (CDRs) that contact cognate antigens.
The variable region is generated stochastically through a process called V(D)J recombination. This process involves the pseudo-random selection, recombination, and joining of gene segments—Variable (V), Diversity (D), and Joining (J) segments—along with the introduction of pseudo-random nucleotide insertions and deletions between the recombined genes. This generates astromomical standing antibody sequence diversity which could never be encoded in the genome. Selection of particular antibodies from this diversity allows indivuduals to adapt their immune system to never-before-seen pathogens.
Antibody Evolution
The antibody generation process creates a large pool standing diversity in protein space. Next, the immune system uses an evolutionary process to select and amplify the antibodies which have the most beneficial properties, such as strongly binding and neutralizing a pathogen which is wreaking havoc in your body. During the selection process, additional diversity is generated by mutating the selected antibodies, allowing the immune system iterate on creating an even better protein: it selects the best of the selected. This process is called somatic hypermuation, and it's pretty much fascinating.